3.3. The i-th tetrahedral alpha-carbon atom
3.3.1.
Vectors analysis of the of the
tetrahedrons formed by the i-th alpha-carbon atom
Bonds of the carbon atom Rai, to which side chains
of a canonical set of amino acids in MVM are attached, as well as other carbon
atoms have a tetrahedral structure, i.e. all four bonds are directed to the
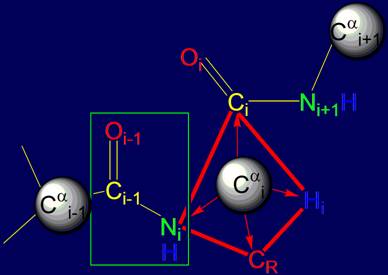
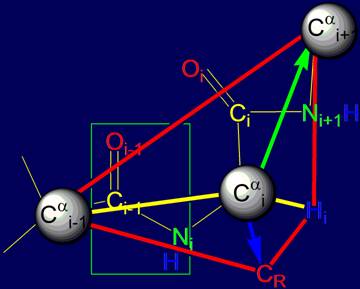
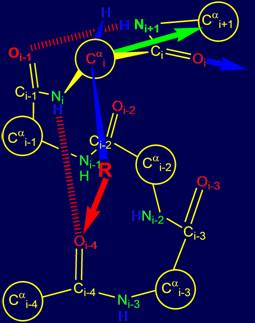
vertices of a tetrahedron. As seen in Figure 14, at these vertices are the
atoms of Ni, Нi, Сi, (group Oi=Ci–Ni+1H) and CR (side chain carbon
atom).
As shown in Fig. 14, a and b, the resonance group Oi-1=Ci-1–NiH
is flat (enclosed in box) [15],
is a single entity and can rotate freely. Through it you can spend a notional axis Cai-1 – Cai,
which will rotate
the atom Cai (Fig.
14,b).
Since all
the bonds of i-th
tetrahedral carbon atom are rigid,
the vectors directed to i+1- th atom of the main chain (Cai+1) and
the side chain atom of СR will
be firmly fixed and interrelated. In Fig.
14,b they are,
respectively, green and blue. This means
that any movement of the side chain of amino acids in the process of reconstruction of the encoded protein conformation would simultaneously affect
the direction of the vector Cai – Cai+1.
For this ability, these two directions at the i-th alpha-carbon atom are called "yoke".
Different
side chains of the protein associated with the tetrahedral a-atom in the
framework of MVM will predetermine the different growth direction of the main chain. Let’s consider this question in more detail.
|
|
|
|
Fig. 14. Comparison of the tetrahedrons formed by Cai: a – by valence bonds Cai, directed to the vertices
of a tetrahedron;
b - by vectors directed to the Cai (green
arrow) of the main chain and side
chain of СR (blue arrow). |
|
3.3.2. The possible bonds
of group Oi=Ci–Ni+1Н with main chain
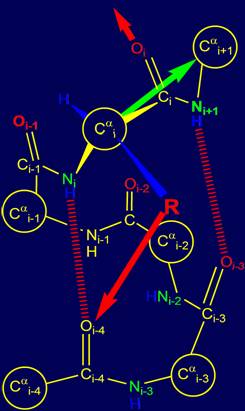
Depending on the
location of bond of group Oi=Ci–Ni+1Н to the main chain it is possible to distinguish four different directions, two of which are shown in Figures
15,a, b.
|
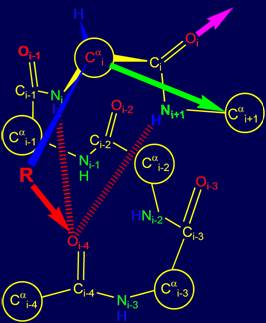
In the event that the
side chain (physical operator) R sets the direction in
which the hydrogen bond between the Ni+1Н and atom Oi-3 is possible (Fig. 15, a), the alpha
helical conformation of the chain [15] remains the same
(recall that previous hydrogen bond was NiН…Oi-4). The direction of the next
hydrogen bond to Oi is shown by
a red arrow. |
|
|
When the physical
operator sets the direction, allowing the formation of hydrogen bonding of
group Oi=Ci–Ni+1Н
and atom Oi-2
(Fig. 15, b), the
spiral structure becomes steeper, which corresponds to the helix 310 of proteins [15]. The direction of the
next hydrogen bond to Oi is shown by a blue arrow. |
|
Fig. 15. Variants of possible hydrogen bonding of Oi=Ri–Ni+1Н: with the atom Oi-3 (a) and with the atom
Oi-2 (b). |
|||
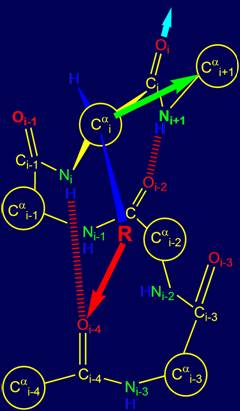
Besides helical structures the side chains,
available in the canonical set, can focus a direction on Сai+1 carbon atom with formation of hydrogen bonds with atoms Oi-1 and on Oi (Fig. 16a, b), which
leads to a steep bends and turns of the chain.
|
The third version of the
bond, which can be defined by physical operators of the i-th position, is formed
by a group of Oi=Ri–Ni+1Н
with the atom Oi-1
Fig. 16, a). It is likely that for
the implementation of this direction is permissible to use only a few side chains
from the canonical set. The direction of the
next possible hydrogen bond for Oi-1 is shown by the blue arrow. |
|
|
The fourth version of the hydrogen bond,
shown in Figure 16,b,
is the formation of hydrogen bonds
with the atom Oi-4.
This corresponds to a situation when with these atoms
are formed simultaneously two hydrogen bonds
- NiH and
Ni+1H by
groups belonging to the i-th and i +1- th alpha-carbon atoms. Such a situation is allowed only for the
amino acids glycine
(Gly), which
has no side chain and having
the conformational mobility. The direction of the next possible hydrogen
bond for the Oi-1 shown
by the pink arrow. |
|
Fig.
16. Variants of
possible hydrogen bonding of group Oi=Ri–Ni+1Н
with atom Oi-1 (a) and with the atom Oi (b). |
|||
Thus, depending on the
length of the side chain, located in the i-th position, determination of the subsequent direction of growth of the polypeptide
chain is possible. Note also that this
determination also depends on the structure of the source pentafragment,
whether it is in the alpha-helical conformation or otherwise.
In this regard, "yoke",
i.e. the relationship of the two directions on the i-th alpha-carbon atom, there is a problem of establishing the correlation between the size
of the side chain and the direction which it determines. For the decision of this question could be used as a
mathematical (i.e., geometric), and computer approaches. The solution of
complex problems in this area, could spell, as it can be assumed to obtain
important results for the direct prediction of protein secondary structure.
Attempts were made in this direction [6-8], but have not yet led
to a complete solution of the problem.
Theoretical analysis of bond
area NiН…Oi-4 and formulated as a result of this representation of the MVM can clarify the
nature of the canonical set
of
20 amino acids as a group of irreducible
representations of vectors that can
be distinguished in this area.
However, this view is
somewhat simplified. As we saw on the page http://genetic-code.narod.ru, in the base of the
genetic code could be laid the 64 conformations of 4-arc graph or protein penafragment,
of which two or three conformations are associated with the encoding of the
beginning or the end of the reading (triplets coding stop-signals). The remaining 61-62 conformations of
the protein must be recreated
by means of side chains. The question
arises: how to reconcile the
existence of only 20 amino acids
with the possibility of coding by
triplets (although highly degenerate) over the 60 conformations of
the protein. This question is
solved in principle, if we
include into consideration not
only the side chains, but also those groups that they carry. The inclusion of these groups leads to a two-layer model
of MVM, which is set out in section 4 of our pages.
Address for connection: vector-machine@narod.ru