3.2. The amino acids
canonical set
On page http://amino-acids-20.narod.ru, dedicated to the spatial
structure of the canonical set of amino acids, it was said that the explanation
of the nature of this set, as well as principles of arrangement of the side
chains of amino acids in the dodecahedron model will be given in model MVM. We
recall these principles:
1. The basis for building -
the structure of the dodecahedron.
2. An arrangement of side
chains on the dodecahedron - in order of increasing of molecular weight.
3. The side chains of amino
acids located on the dodecahedron with respect to each other on the basis of
antisymmetry principles.
We offer such an explanation, using
group-theoretic approach used in Section 3.1. for the analysis of the vectors
of action.
From the definition of representations of the group [16] follows. Suppose there is a finite group G with elements g1,
g2, … gn. If the
group T of linear operators Tgi in some space R homomorphic to group G, we say
that the group T forms a representation of group G. From the analysis of
properties of linear operators, it follows that each element of gi
group can be associated with the matrix || Drk(gi)||. In
this case the neutral element must be mapped to a neutral matrix and the
inverse element - the inverse matrix.
By analogy with the elements of
group and their representations, the side chains of amino acids, regarded by us
as physical operators can be called "irreducible representations" of
the group of vectors. The definition of "irreducible" is used by us
in the sense that these side chains are the most simple and cannot be easier.
Using this analogy, we associate a vectors groups and groups of amino acid side
chains (physical operators), recreating the effect of these vectors. This
comparison is useful in the development of ideas about the structure, which
should have a canonical set of side chains.
3.2.1. The side chains of amino acids realizing action of neutral and inverse
elements
Let's carry out the analysis of some amino acids as the elements of
group of irreducible representations. In this group, starting from the axioms
of mathematical group should be the elements realizing action the action of
neutral and inverse elements.
Vector, which is a neutral element in group of vectors must clearly
recreated by the physical operator, which will not influence the structure of
the protein pentafragment. This means that such an operator may have no side chain, and be
"empty" link in the chain of the protein. According to its properties
it should refers to a subgroup of operators of connectivity. The only amino
acid that is suitable for this role is glycine (Gly), which has no side chain,
and in fact is "empty" link in the chain of the protein (the structure
of Gly and of other amino acids see on the http://amino-acids-20.narod.ru/aminoacids.htm).
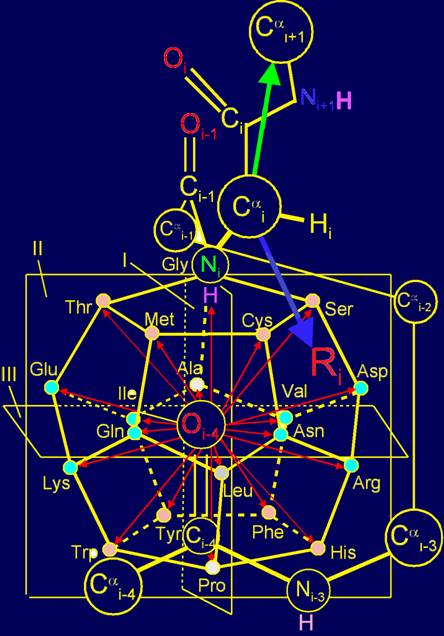
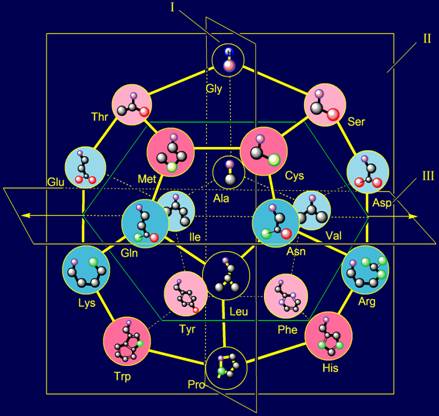
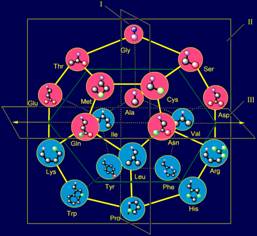
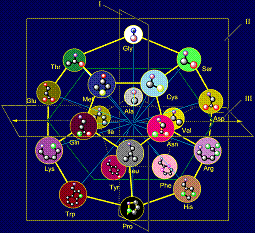
In model of structure of the canonical set of amino acids on the
dodecahedron (Fig. 11, b) Gly is located in the plane of I and is one of the
top vertices. In the MVM (Fig. 11, a) its nitrogen atom Ni atom becomes associated with the Сai-atom of backbone. The absence of side chain in this amino acid leads to
the fact, that the connection between NiH…Oi-4=C is unregulated. As you know, Gly has a
high conformational mobility [15], which contributes
to the function of glycine as neutral element.
|
|
|
|
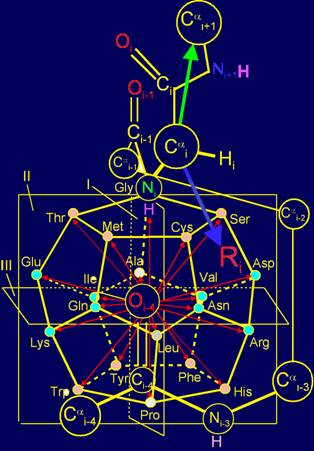
Fig. 11. Molecular Vector Machine of proteins (a), and the position of
amino acids realizing the action of neutral and inverse elements (b) directed
along the bond NiH…Oi-4=C, as well as of amino acids,
realizing the action of vectors directed across this link (c) in the model on
the dodecahedron. |
|
Vector corresponding to the inverse element of group has an opposite
direction by the action, directed on rupture of bond NiH…Oi-4=C. It can realized by a physical operator,
whose Ni atom, in principle, cannot form a hydrogen bond with the group Oi-4=C. Such a situation is possible, for
example, if the Ni atom is included in a cyclic structure. At the same atom Oi-4
can purchase the opposite direction of the hydrogen bond as a result of
"front" collision of its electronic cover with the electronic cover
of these non polar cyclic structures.
The most suitable for the role of the inverse element of the side chain
is proline (Pro). Take a look at its cyclic structure: http://amino-acids-20.narod.ru/aminoacids.htm. This is the only side chain, nitrogen atom of which forming a peptide
bond in the main chain, is included in a cycle. This atom cannot in principle
form a hydrogen bond with the group Oi-4=C with the formation of cyclic
pentafragment, cyclic part of the proline may be involved in a
"front" collision with this group, and thus realize the effect of the
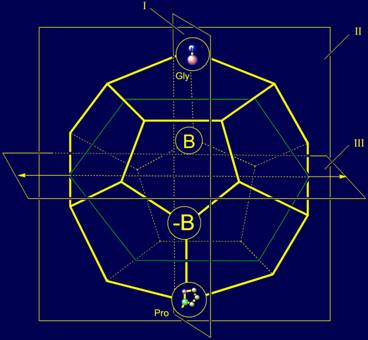
vector, designed to break the H bond. In Figure 10, b, in the model of
structure of the canonical set of amino acids in the dodecahedron proline is
also located in the plane of I, but in one of the lower vertices.
Thus, in the canonical set of amino acids side chains there are two side
chains - Gly and Pro, which can serve as, respectively, neutral and reverse
elements when considering them as irreducible representations of vectors.
In addition to the action along the bond NiH…Oi-4=C, a group of vectors has a subgroup whose
action is directed across this bond. They also located in the plane I (see Fig.
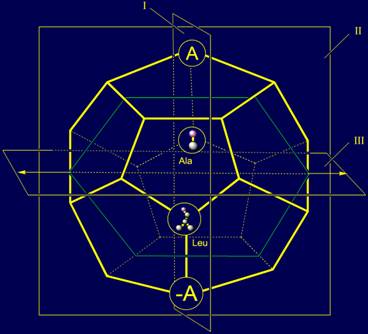
11, a). On the role of representations of these vectors two amino acids -
alanine (Ala) and leucine (Leu) are suitable. Their position in the model of
structure of the canonical set of amino acids on the dodecahedron is shown in
Figure 11, c.
3.2.2. Arrangement of amino acids
in ascending order of molecular weight
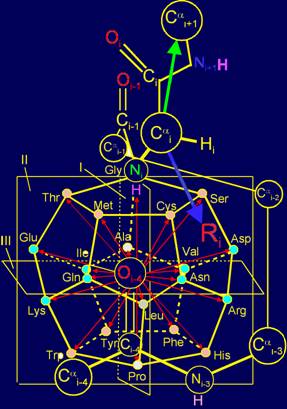
As can be seen from Figure 12,a
the point of application of the set of vectors (red arrows) and a variety of
side-chain amino acids (blue arrow), denoted as Ri, are dodecahedron vertices. On the one
hand, these are the points at which are directed vectors acting on the link NiH...Oi-4=C, and on the other hand, they are the place
of application of side chains of amino acids in the role of physical operators
realizing action of these vectors.
Vector in МVМ have a various direction: a part of them
is directed towards atom Ni, and a part - to an opposite side. Accordingly, the side chains of the
amino acids, recreating vectors directed towards atom Ni as can be seen from Figure 12,a
should be shorter than side chains, recreating the vector directed toward the Оi-4=С.
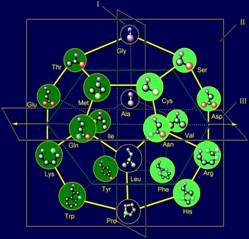
The side chains of amino acids in the model on dodecahedron (Fig. 12, b)
are arranged in order of increasing of their mass and, consequently, the size
that has provided realization of this principle in МVМ.
Shorter side chains (Ser, Thr, Cys, Met - in pink circles), as shown
Figure 12, b, located at the top of the dodecahedron, and will recreate the
vectors directed along the plane of I in the direction of atom Ni. More massive, such
as cyclic (Phe, Tyr, His, Trp, also in pink circles) are located at the bottom
and will re-create the vectors, also directed along the plane of I, but in the
opposite direction.
In the center of the dodecahedron are the side chains, recreating the
vectors directed along the plane III – these are the side chains of average
size and weight (Asp, Asn, Arg, Glu, Gln, Lys - in blue circles). Details of
the location of the side chains on the dodecahedron are stated on a site http://amino-acids-20.narod.ru/AA_dodecahedron.htm.
|
|
|
|
Fig. 12. Molecular Vector Machine of proteins (a) and model of the
structure of the canonical set of amino acids on the dodecahedron (b). |
|
Thus, the principle of an arrangement of the side chains of amino acids on
the dodecahedron in order of increasing size and molecular weight is caused by
that the side chains, while increasing size, provide realization of action of
all directions of the vectors, set by structure of the dodecahedron.
3.2.3. Antisymmetry of the side chains
As on model structure of the canonical set of amino acids, there are
three planes of antisymmetry, transition through these planes to antisymmetric
amino acid can be designated by any letter. On page http://amino-acids-20.narod.ru/AA_antisymmetries.htm
following designations have been entered:
- Amino acid transition in itself is designated by figure 1.
- Transition through a plane I - the letter a (alpha),
- Transition through a plane II - the letter b (beta);
- Rotation round an axis laying in a plane III - the letter g (gamma).
Then all transformations of the amino acids located in a column 1, in
each group may be described in the form of the following table.
|
|
1 |
a |
b |
g |
ab |
ag |
bg |
abg |
|
Subgroup 1 |
Gly |
|
|
Pro |
|
|
|
|
|
Subgroup
2 |
Ala |
|
|
Leu |
|
|
|
|
|
Subgroup
3 |
Ser |
Thr |
Cys |
His |
Met |
Trp |
Phe |
Tyr |
|
Subgroup
4 |
Asp |
Glu |
Asn |
Arg |
Gln |
Lys |
Val |
Ile |
These antisymmetric transformations in principle repeat the symmetrical
transformation of vectors, which recreate the side chains of amino acids (Section
3.1.), but instead of the symmetry for the side
chains of amino acids appears antisymmetry.
On page http://amino-acids-20.narod.ru/AA_antisymmetries.htm it is shown that the structure of the dodecahedron reveals four types
of antisymmetry available to the side chains of amino acids: a
quasi-mirror-like, non-mirror, rotating, and complementary. Using the approach
to the side chains as to the irreducible representations of the vectors allows
a unified explanation to the nature of these types of antisymmetry.
|
Quasi-mirror |
Non-mirror |
||
|
The side chains of amino acids,
recreating the vectors directed to the right of the plane I, should be
shorter. Position of Сai -atom to which side chains of amino acids as physical operators of
protein MVM are attached, as seen in Figure 13,a is a little asymmetric and
shifted to the right from the link NiH...Oi-4=C. This means that the side chains that recreate
symmetrical with respect to the plane I vector directed to the right of this
plane should be shorter than the side chains, realizing the action of vectors
directed to the left of this plane. This pattern should appear on all side chains, which should lead to
the formation from them two groups of amino acids having similar properties
but different lengths. That is why the canonical set of amino acids consists
of two groups that could be arranged on the dodecahedron symmetrically about
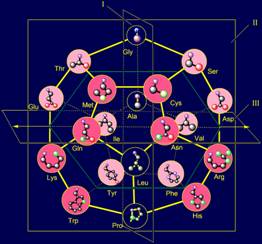
the plane I (Fig. 13,b). This corresponds to a quasi-mirror
antisymmetry: (http://amino-acids-20.narod.ru/quasi_mirror_antisymmetry_AA.htm). |
Similar in size, but different in
properties, the side chains should recreate vectors which are arranged
symmetrically with respect to the plane II.
As can be seen at Figure 13,a, the distance from the Сai-atom to the vertices, to which symmetric vectors are directed being behind
the plane II, (are directed from us), and in front of the plane II (are
directed to us) differ not so strongly. For this reason, the side chains
recreating these vectors should not vary significantly in size. These
differences can only consist in the replacement of terminal groups. Indeed,
pairs of amino acids located in the plane II and behind it are close (Fig.
13,c). This corresponds to their arrangement on the basis of a non-mirror antisymmetry: ( http://amino-acids-20.narod.ru/non_mirror_antisymmetry_AA.htm ). |
||
|
|
|||
|
Fig. 13. Molecular Vector Machine of proteins (a) and types of
antisymmetry, revealed by the model structure of the canonical set of amino
acids on the dodecahedron: a quasi-mirror (b), a non-mirror (c), rotational
(d) and komplementary (e). |
|||
|
Rotational |
Комплементарная |
||
|
Shorter side chains of amino acids
recreate vectors, located above the plane III. As
seen in Figure 13,a, the vertices of the vectors directed towards the atom NiH, located above the plane III, are much
closer to the Sai-atom than of ones symmetric to them, but aimed in the
opposite direction to the atom Oi-4 (located below the plane III .) For this reason, all the vectors that
are above the plane III, should be recreated by the shorter side chains of
amino acids (physical operators) than under the plane III. Since these
vectors are connected by operation of rotation round the axis C2 (g), amino acids with the shortest side
chains should withstand the longest. At the same time, the side chains, recreating the vectors located in
the equatorial region of the dodecahedron must differ to a lesser extent, for
example, one link in the chain. That is what we observe in the structure of
the canonical set of amino acids on the dodecahedron (Fig. 13, d). This
corresponds to a rotational antisymmetry of the side chains of amino acids: (http://amino-acids-20.narod.ru/rotary_antisymmetry_AA.htm). It becomes clear why there is such antisymmetry in a canonical set of
the side chains of amino acids. |
The side chains that recreate the vectors
of opposite direction should have the additional properties. This is due to the fact that the slope of the axes of these vectors is
the same, together both vectors make up its diameter (see Fig. 13, a). In
this case, since the diameters are connecting the vertices of the
dodecahedron, located on opposite sides of the plane I, the side chains which
are on one axis and situated to the right of the plane I will be maximally
close to Сai -atom, and to the
left of the plane - maximally far away from Сai-atom. This allows us to understand why the structure of amino acids
(Fig. 13, d) at the vertices of the dodecahedron, connected by diameters are
the side chains, which properties are additional (complementary) - antisymmetry of complementarity:
(http://amino-acids-20.narod.ru/AA_complementarity_antisymmetry.htm) . The shortest chain of Ser linked to the most massive - Trp, the
smaller, compared with Trp His chain is associated with more massive compared
to Ser, a chain of Thr. The minimum sized negatively charged side chain of
Asp is on the same diameter with a maximum length positively charged chain of
Lys, but a shorter chain of Arg is associated with a bigger in length chain
of Glu, etc. The direction of Сai – Сai+1 given by each of the side chains, recreating the vectors of the same
slope, should also, apparently, be the same. |
||
Thus, the existence of four types of anti-symmetry, revealed by the model
of structure of the canonical set of amino acids on the dodecahedron, is due to
the participation of the side chains of amino acids in the realization of the
symmetric vectors of action with a different direction in MVM.
The next part of the MVM, discussed in Section 3.3., is the i-th tetrahedral
alpha-carbon atom, which serves as a place of an attachment of exchangeable
side chains of amino acids of the canonical set.
Address for connection: vector-machine@narod.ru